ASP 460 2.0 Data Visualization
Dr Thiyanga Talagala
Visualizing Time Series Data
Individual Time Series: Points
library(mozzie)library(ggplot2)data(mozzie)colombo.dengue <- mozzie[, 1:4]ggplot(colombo.dengue, aes(x=ID, y=Colombo))+ geom_point()+xlab("Time")+ ylab("Number of dengue cases")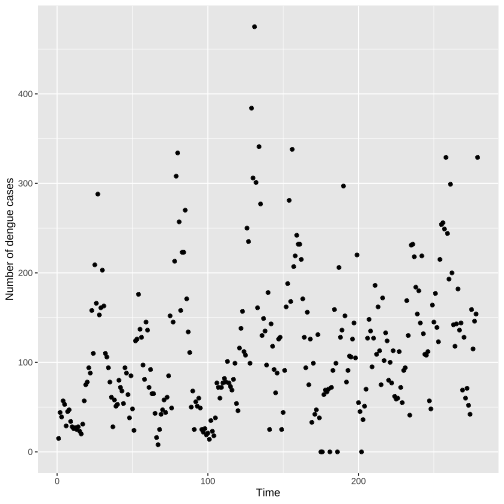
Individual Time Series: Points
library(mozzie)library(ggplot2)data(mozzie)colombo.dengue <- mozzie[, 1:4]ggplot(colombo.dengue, aes(x=ID, y=Colombo))+ geom_point()+xlab("Time")+ ylab("Number of dengue cases")This is NOT a scatter plot. Why?
Individual Time Series: Points
This is NOT a scatter plot. Why?
the points are spaced equally along the x-axis.
there is a order among points.
To emphasize time dependent relationship we can connect neighboring points with lines.
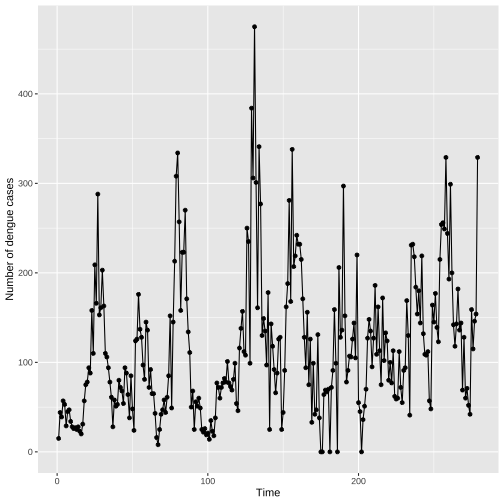
Individual Time Series: Points and Lines
library(mozzie)library(ggplot2)data(mozzie)colombo.dengue <- mozzie[, 1:4]ggplot(colombo.dengue, aes(x=ID, y=Colombo))+ geom_point()+ geom_line()+ xlab("Time")+ ylab("Number of dengue cases")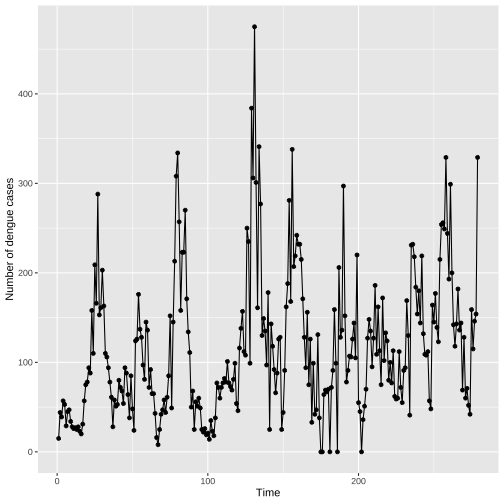
Individual Time Series: Points and Lines
Lines do not represent observed data. Lines are meant as a guide to the eye.
If few observed values a far apart or unevenly spaced, it is not suitable to connect points with lines.
Individual Time Series: Lines only
ggplot(colombo.dengue, aes(x=ID, y=Colombo))+ geom_line()+ xlab("Time")+ ylab("Number of dengue cases")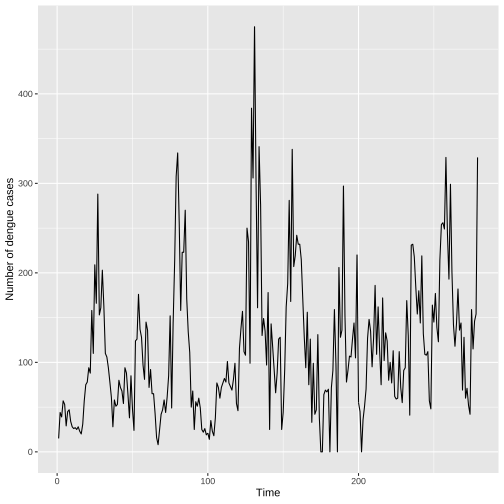
Individual Time Series: Lines only
Without points more emphasis is given on the overall trend and less on individual values.
In general, when there are too many points it is better to plot without points.
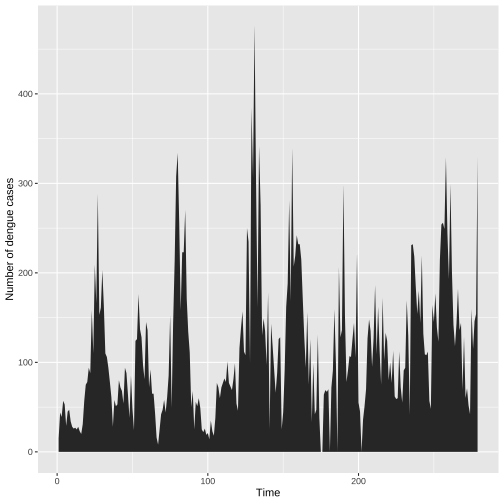
Individual time series: Fill the area under the curve
ggplot(colombo.dengue, aes(x=ID, y=Colombo))+ geom_area()+ xlab("Time")+ ylab("Number of dengue cases")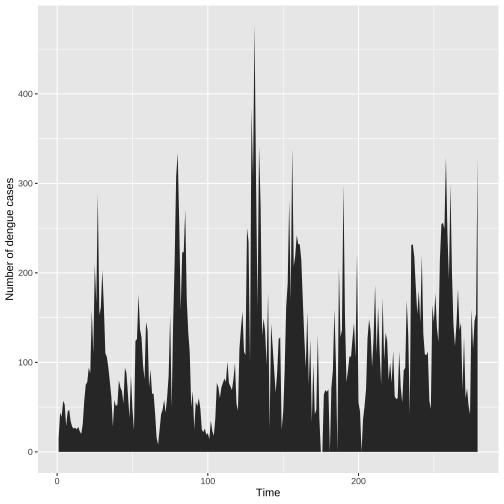
Visually separates the area above and below the curve.
More emphasis is given to the overarching trend in the series.
This visualization is only valid if the y axis starts at zero.
Visualising multiple time series
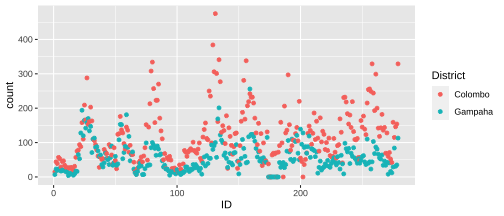
Difficult to read.
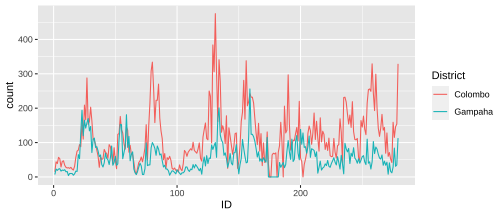
By connecting points with lines we help the reader to follow the paths of each individual time series.
Smoothing
library(gapminder)head(gapminder)## # A tibble: 6 x 6## country continent year lifeExp pop gdpPercap## <fct> <fct> <int> <dbl> <int> <dbl>## 1 Afghanistan Asia 1952 28.8 8425333 779.## 2 Afghanistan Asia 1957 30.3 9240934 821.## 3 Afghanistan Asia 1962 32.0 10267083 853.## 4 Afghanistan Asia 1967 34.0 11537966 836.## 5 Afghanistan Asia 1972 36.1 13079460 740.## 6 Afghanistan Asia 1977 38.4 14880372 786.Smoothing
library(tidyverse)gapminder_af <- gapminder %>% filter(continent == "Africa")ggplot(gapminder_af, aes(x=year, y=lifeExp))+ geom_point()+ geom_line()+ facet_wrap(~country)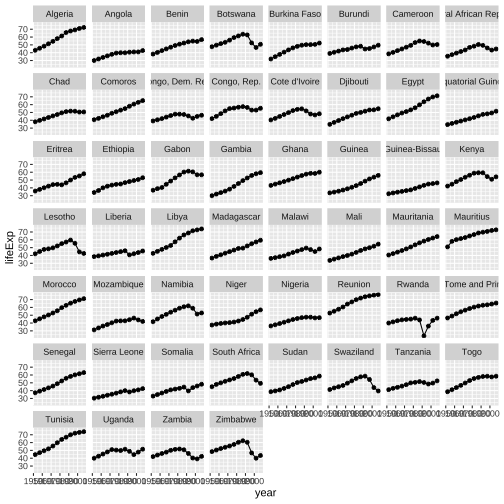
Smoothing
gapminder_af <- gapminder %>% filter(continent == "Africa")ggplot(gapminder_af, aes(x=year, y=lifeExp))+ geom_line()+ facet_wrap(~country)+ theme(axis.text.x = element_text(angle = 90, hjust = 1))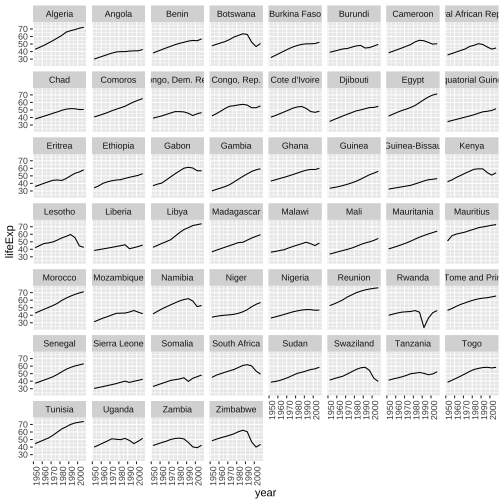
Smoothing
library(tidyverse)gapminder_af <- gapminder %>% filter(continent == "Africa")ggplot(gapminder_af, aes(x=year, y=lifeExp))+geom_smooth()+facet_wrap(~country)+ theme(axis.text.x = element_text(angle = 90, hjust = 1))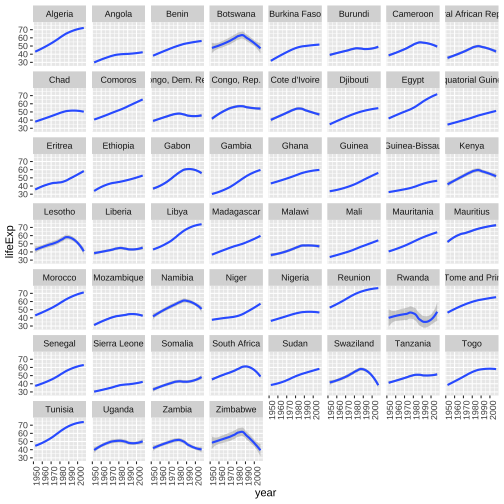
Time series plots
library(forecast)library(fpp2)mozcol <- ts(mozzie$Colombo, frequency = 52, start = c(2008, 52))autoplot(mozcol) + ggtitle("Dengue Count - Colombo") + ylab("Count") + xlab("Year")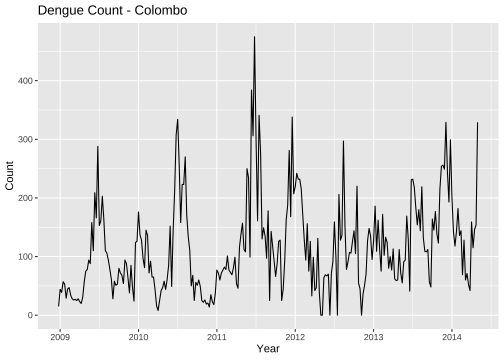
Seasonal plots
ggseasonplot(mozcol, year.labels=TRUE, year.labels.left=TRUE) + ylab("Count") + ggtitle("Seasonal plot: Dengue Count - Colombo")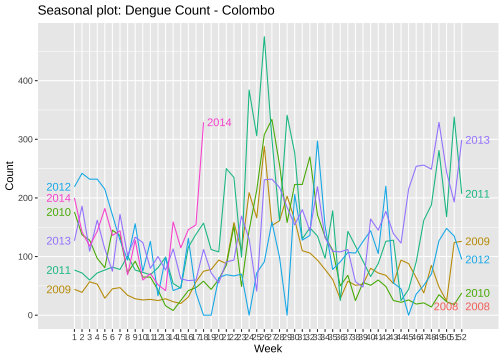
Polar seasonal plot
ggseasonplot(mozcol, polar=TRUE) + ylab("Count") + ggtitle("Polar seasonal plot: Dengue Count - Colombo")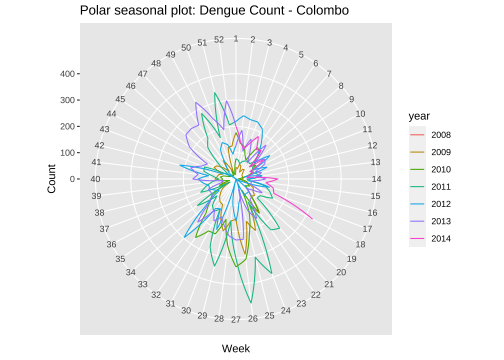
Time series plot
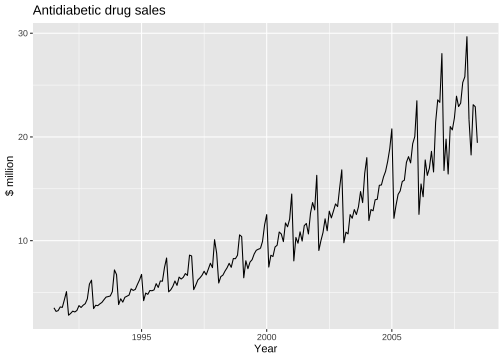
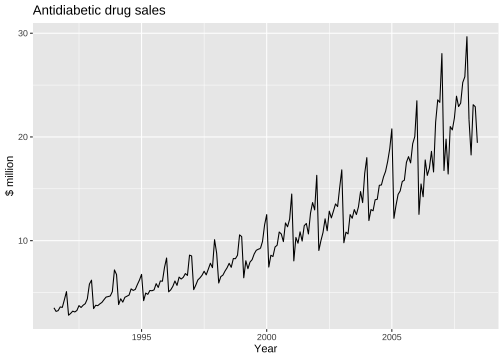
Seasonal plot
ggseasonplot(a10, year.labels=TRUE, year.labels.left=TRUE) + ylab("$ million") + ggtitle("Seasonal plot: antidiabetic drug sales")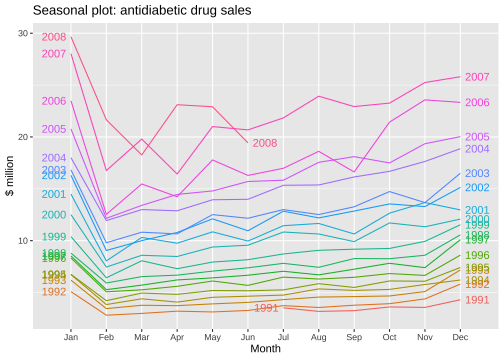
Polar seasonal plot
ggseasonplot(a10, polar=TRUE) + ylab("$ million") + ggtitle("Polar seasonal plot: antidiabetic drug sales")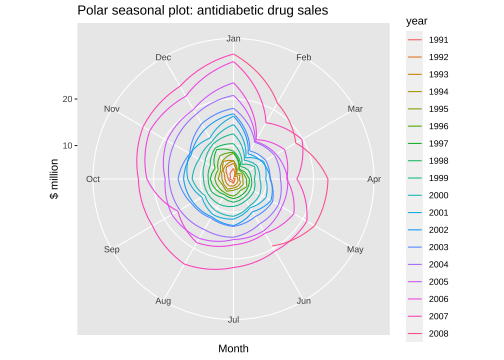
Seasonal subseries plots
ggsubseriesplot(a10) + ylab("$ million") + ggtitle("Seasonal subseries plot: antidiabetic drug sales")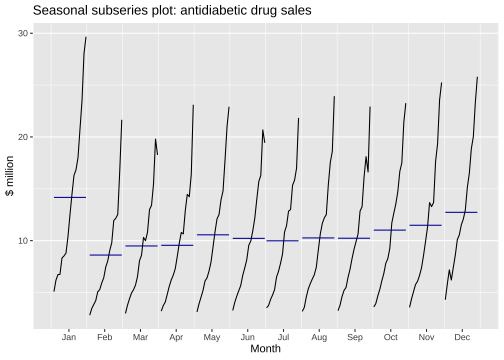
a10 Jan Feb Mar Apr May Jun Jul1991 3.5265911992 5.088335 2.814520 2.985811 3.204780 3.127578 3.270523 3.7378511993 6.192068 3.450857 3.772307 3.734303 3.905399 4.049687 4.3155661994 6.731473 3.841278 4.394076 4.075341 4.540645 4.645615 4.7526071995 6.749484 4.216067 4.949349 4.823045 5.194754 5.170787 5.2567421996 8.329452 5.069796 5.262557 5.597126 6.110296 5.689161 6.4868491997 8.524471 5.277918 5.714303 6.214529 6.411929 6.667716 7.0508311998 8.798513 5.918261 6.534493 6.675736 7.064201 7.383381 7.8134961999 10.391416 6.421535 8.062619 7.297739 7.936916 8.165323 8.7174202000 12.511462 7.457199 8.591191 8.474000 9.386803 9.560399 10.8342952001 14.497581 8.049275 10.312891 9.753358 10.850382 9.961719 11.4436012002 16.300269 9.053485 10.002449 10.788750 12.106705 10.954101 12.8445662003 16.828350 9.800215 10.816994 10.654223 12.512323 12.161210 12.9980462004 18.003768 11.938030 12.997900 12.882645 13.943447 13.989472 15.3390972005 20.778723 12.154552 13.402392 14.459239 14.795102 15.705248 15.8295502006 23.486694 12.536987 15.467018 14.233539 17.783058 16.291602 16.9802822007 28.038383 16.763869 19.792754 16.427305 21.000742 20.681002 21.8348902008 29.665356 21.654285 18.264945 23.107677 22.912510 19.431740 Aug Sep Oct Nov Dec1991 3.180891 3.252221 3.611003 3.565869 4.3063711992 3.558776 3.777202 3.924490 4.386531 5.8105491993 4.562185 4.608662 4.667851 5.093841 7.1799621994 5.350605 5.204455 5.301651 5.773742 6.2045931995 5.855277 5.490729 6.115293 6.088473 7.4165981996 6.300569 6.467476 6.828629 6.649078 8.6069371997 6.704919 7.250988 7.819733 7.398101 10.0962331998 7.431892 8.275117 8.260441 8.596156 10.5589391999 9.070964 9.177113 9.251887 9.933136 11.5329742000 10.643751 9.908162 11.710041 11.340151 12.0791322001 11.659239 10.647060 12.652134 13.674466 12.9657352002 12.196500 12.854748 13.542004 13.287640 15.1349182003 12.517276 13.268658 14.733622 13.669382 16.5039662004 15.370764 16.142005 16.685754 17.636728 18.8693252005 17.554701 18.100864 17.496668 19.347265 20.0312912006 18.612189 16.623343 21.430241 23.575517 23.3342062007 23.930204 22.930357 23.263340 25.250030 25.8060902008Seasonal subseries plots
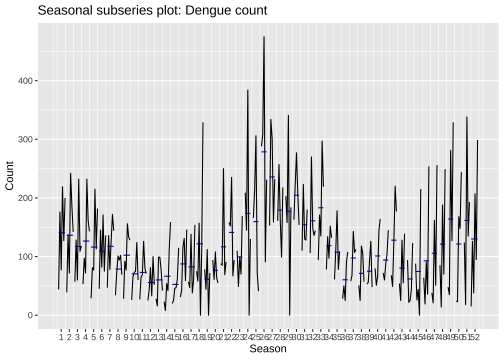
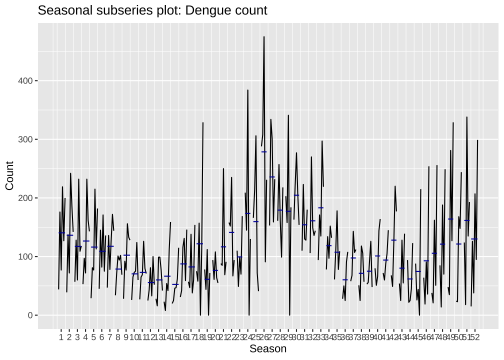
Lag plots: a10
Monthly anti-diabetic drug sales in Australia from 1991 to 2008.
gglagplot(a10)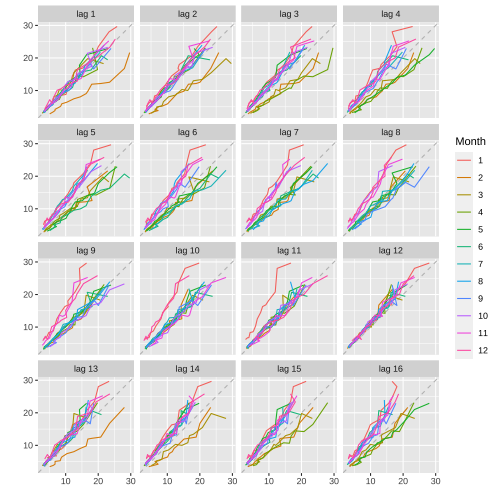
Lag plots: mozzie
Dengue counts - Colombo
gglagplot(mozcol)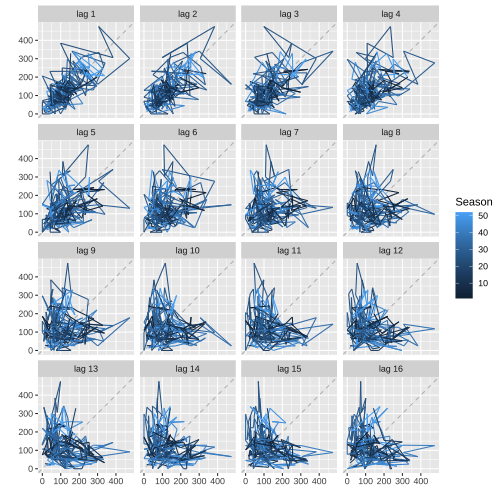
Lag plots: ausbeer
Monthly Australian beer production: Jan 1991 – Aug 1995.
beer2 <- window(ausbeer, start=1992)gglagplot(beer2)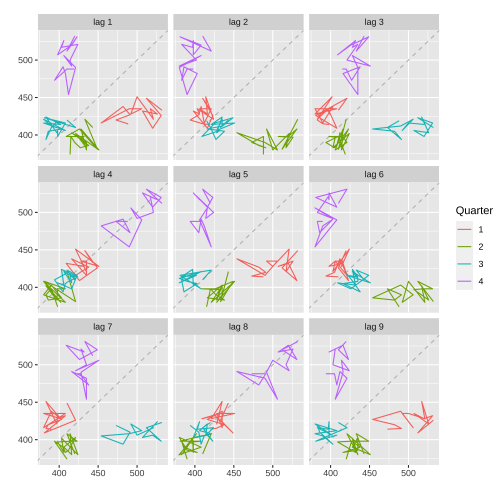
Time series features
Transform a given time series y={y1,y2,⋯,yn} to a feature vector F=(f1(y),f2(y),⋯,fp(y))′.
Examples of time series features
strength of trend
strength of seasonality
lag-1 autocorrelation
spectral entropy
proportion of zeros
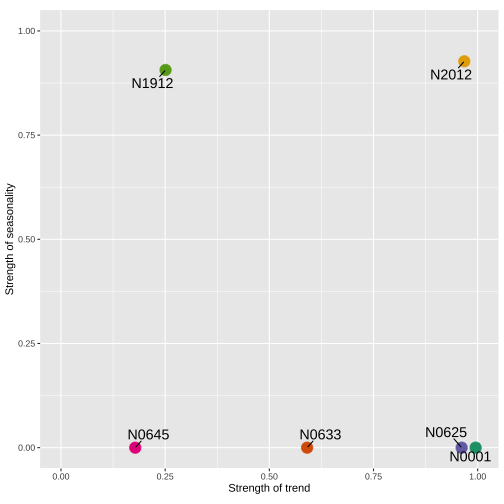
Time-domain representation
Feature-domain representation
References
Talagala, T. S., Hyndman, R. J., & Athanasopoulos, G. (2018). Meta-learning how to forecast time series. Monash Econometrics and Business Statistics Working Papers, 6, 18.