ASP 460 2.0 Data Visualization
Dr Thiyanga Talagala
Colour Scales
1 / 39
Color Scales
Qualitative color scale
Sequential color scale
Diverging color scale
2 / 39
Qualitative color scale
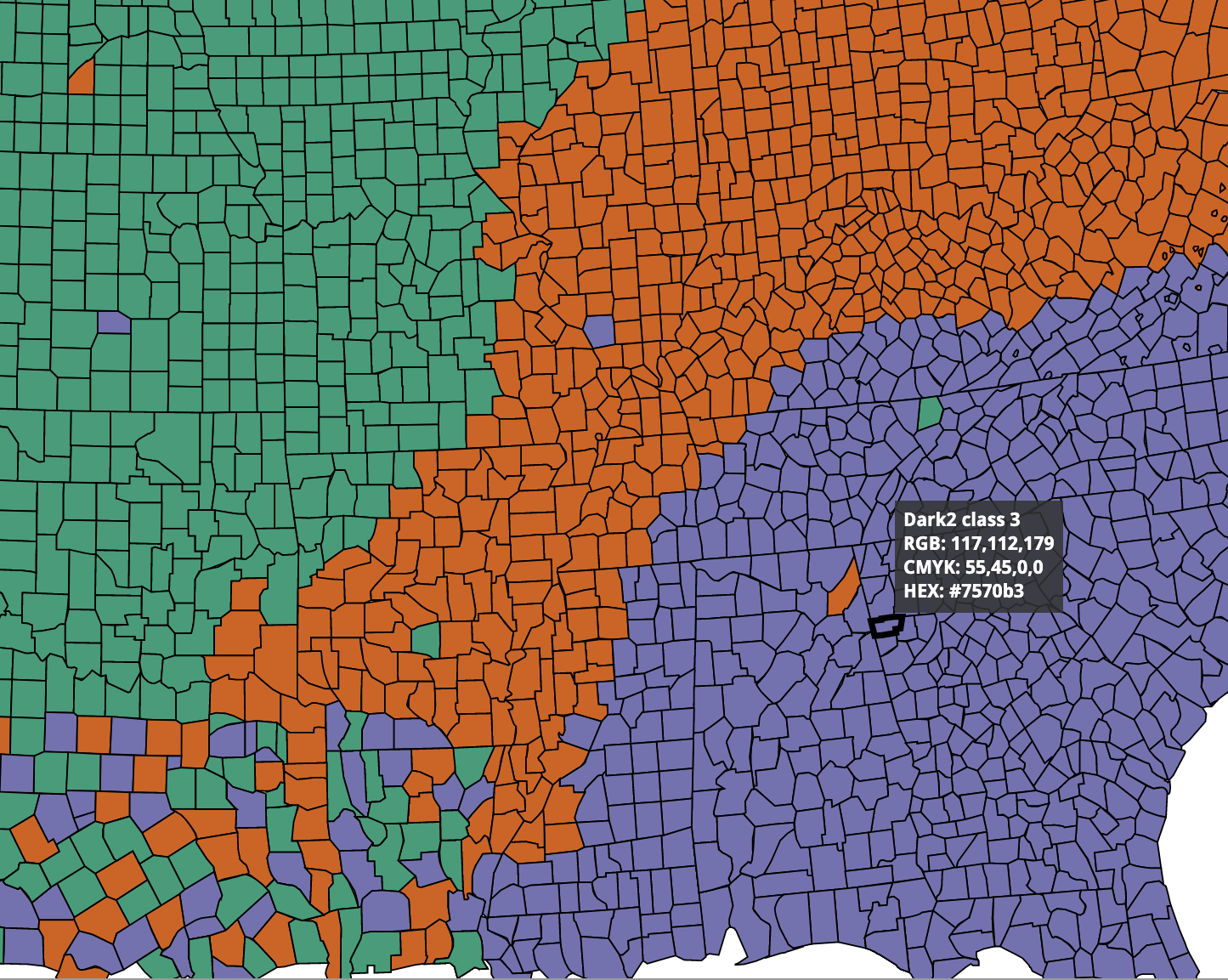
3 / 39
Sequential color scale
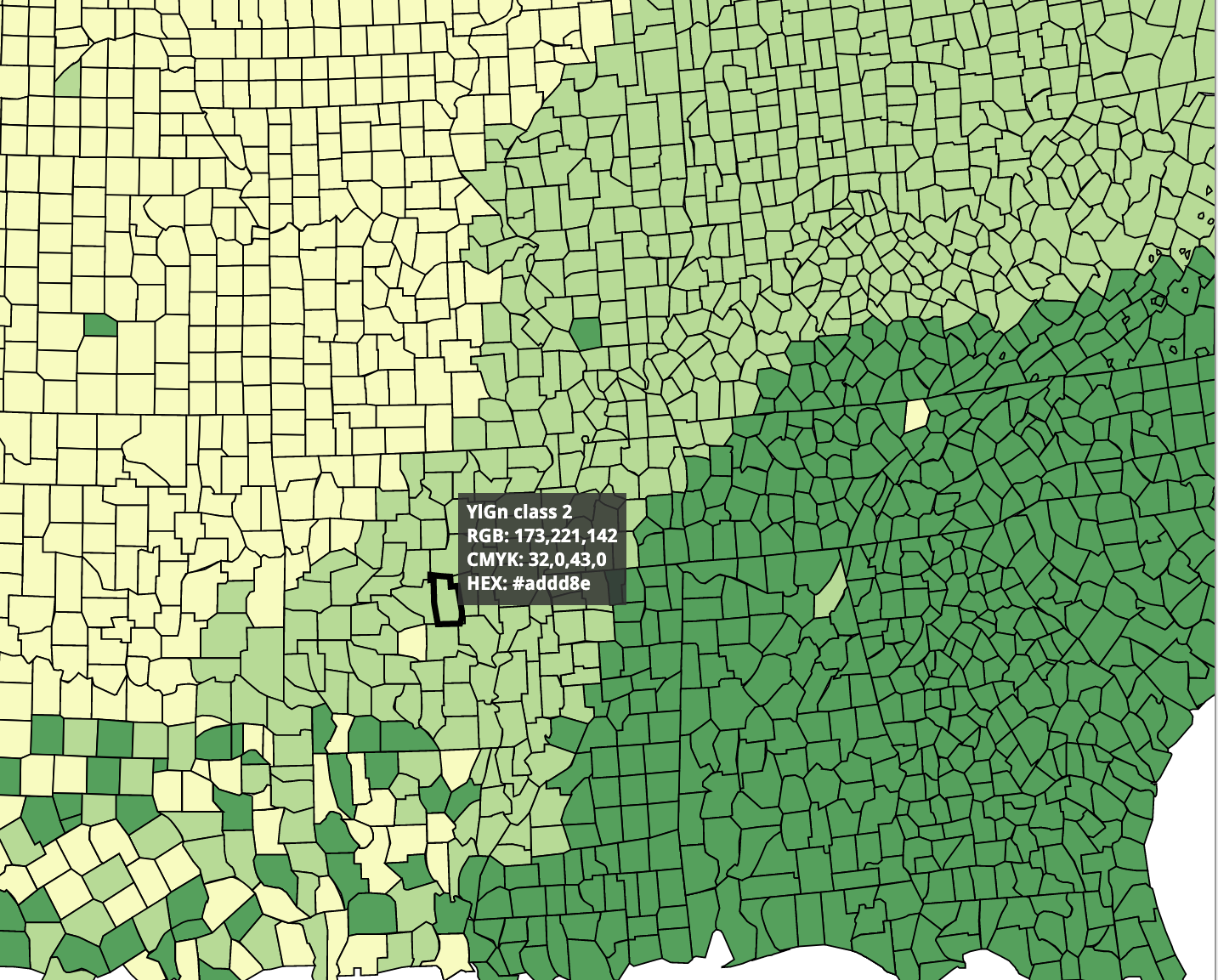
4 / 39
Diverging color scale
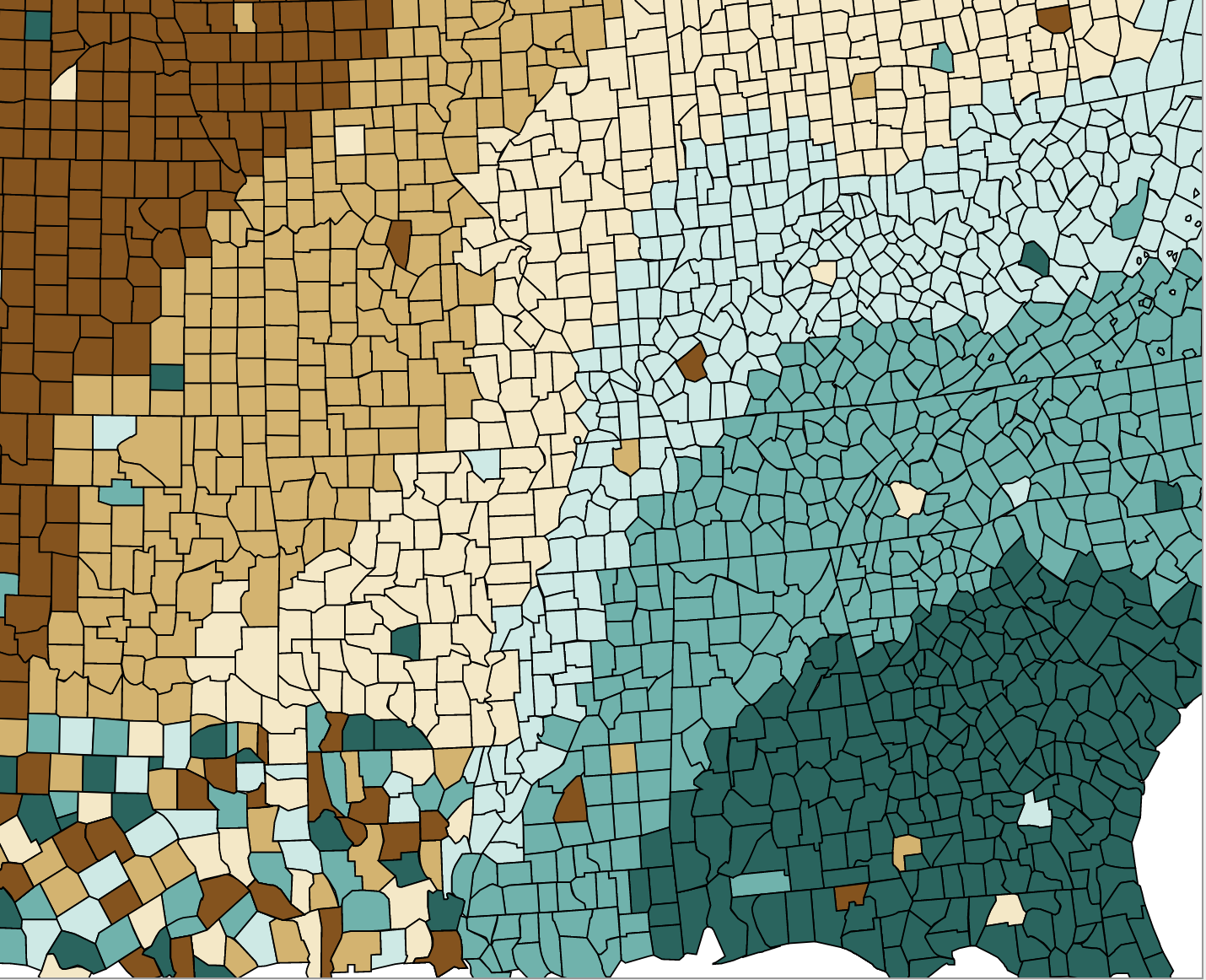
5 / 39
6 / 39
7 / 39
Setting colours in ggplot
8 / 39
library(ggplot2)ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width)) + geom_point()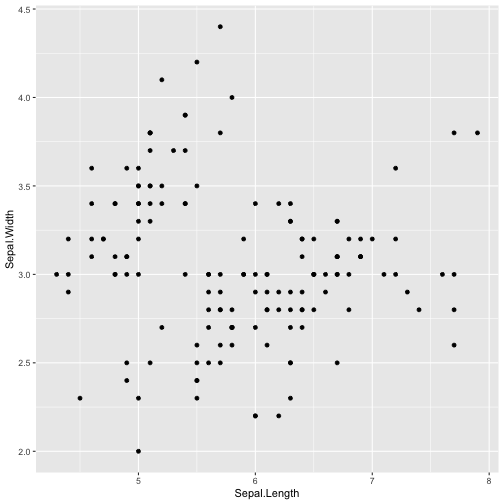
9 / 39
library(ggplot2)ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width)) + geom_point(color="purple")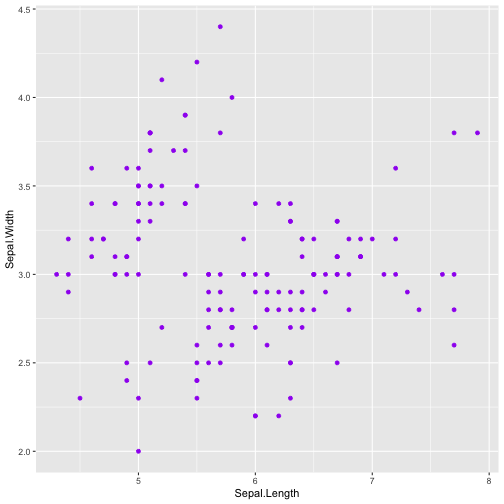
10 / 39
library(ggplot2)ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width, color=Species)) + geom_point()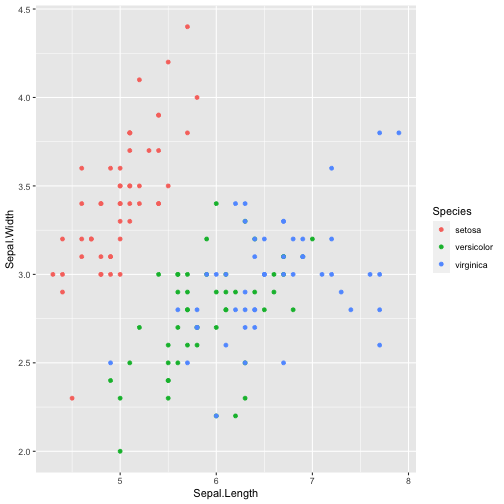
11 / 39
library(ggplot2)ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width, color=Petal.Length < 5)) + geom_point()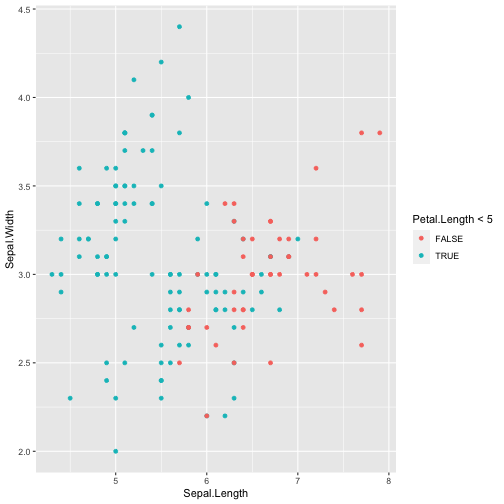
12 / 39
cols <- c("setosa" = "red", "versicolor" = "blue", "virginica" = "darkgreen")ggplot(iris, aes(x = Sepal.Length, y = Sepal.Width, color = Species)) + geom_point( ) + scale_colour_manual(values = cols)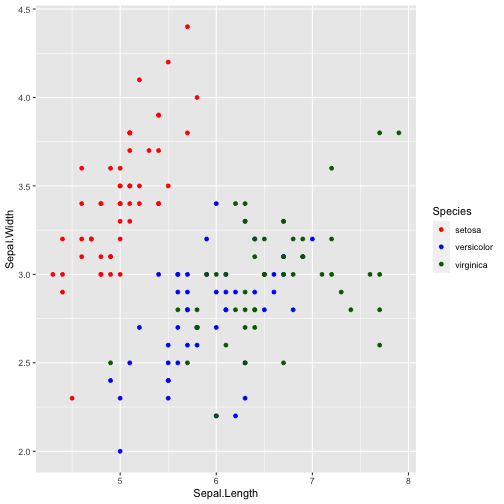
13 / 39
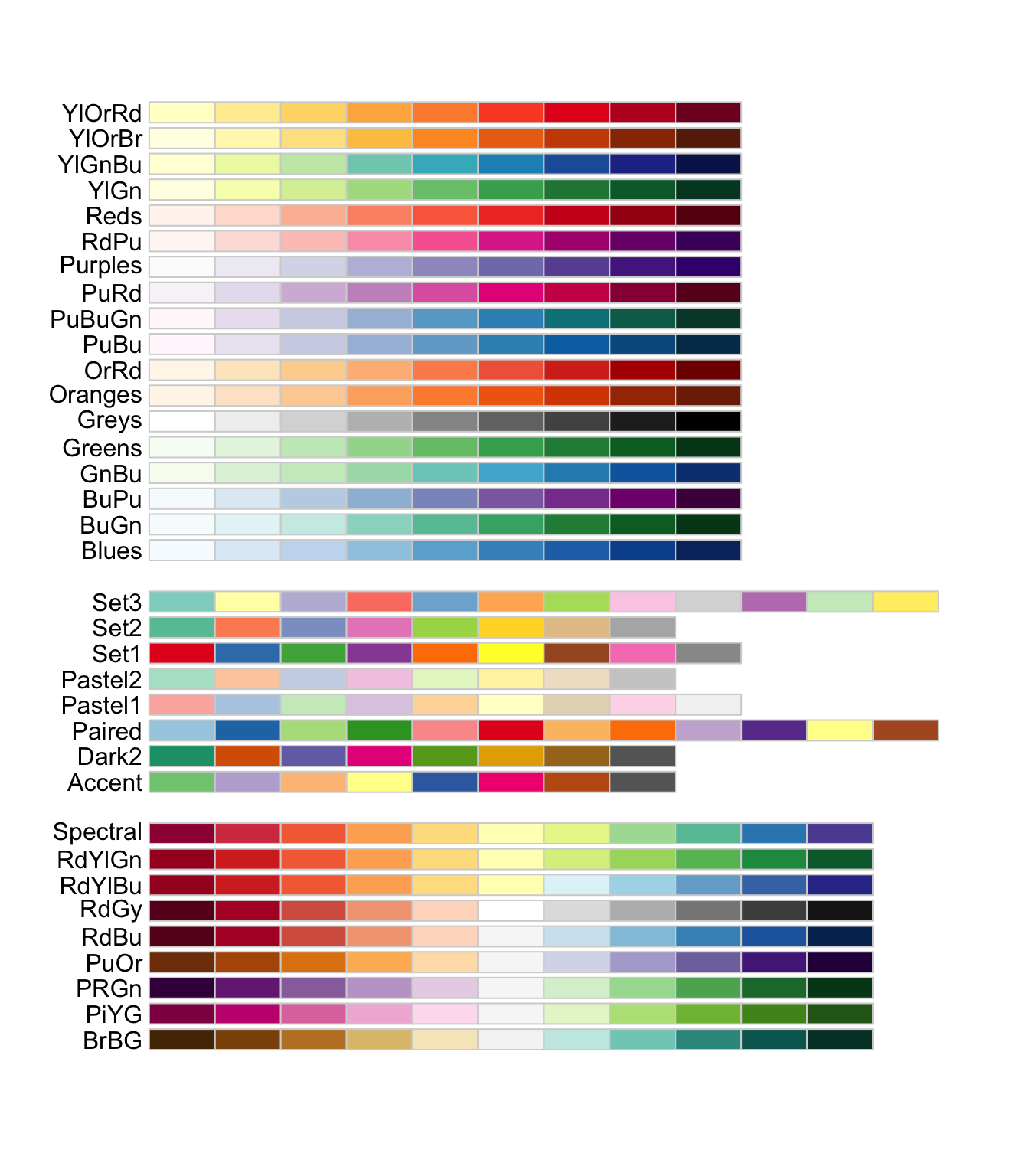
RColorBrewer::display.brewer.all()seq (sequential)
div (diverging)
qual (qualitative)
14 / 39
cols <- c("setosa" = "red", "versicolor" = "blue", "virginica" = "darkgreen")ggplot(iris, aes(x = Sepal.Length, y = Sepal.Width, color = Species)) + geom_point( ) + scale_color_brewer(type = 'qual', palette = 'Dark2')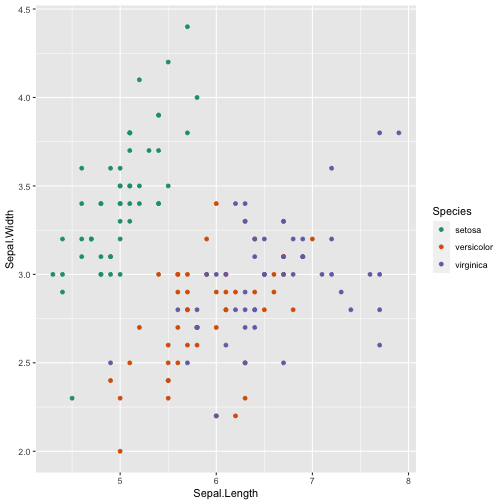
15 / 39
16 / 39
ggplot(iris, aes(x = Sepal.Length, y = Sepal.Width, color = Species)) + geom_point( ) + scale_color_manual(values = c("#1b9e77", "#d95f02", "#7570b3"))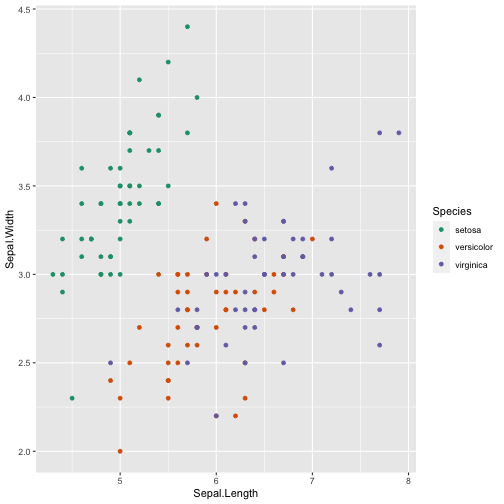
17 / 39
Viridis colour scales from viridisLite
"The viridis scales provide colour maps that are perceptually uniform in both colour and black-and-white. They are also designed to be perceived by viewers with common forms of colour blindness. See also https://bids.github.io/colormap/."
source: https://ggplot2.tidyverse.org/reference/scale_viridis.html
18 / 39
ggplot(iris, aes(x = Sepal.Length, y = Sepal.Width, color = Species)) + geom_point( ) + scale_colour_viridis_d()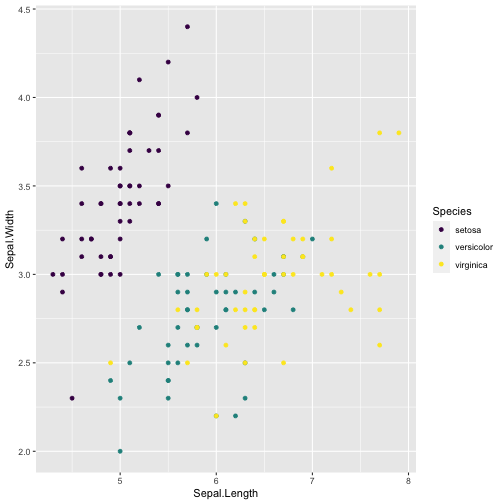
19 / 39
ggplot(iris, aes(x = Sepal.Length, y = Sepal.Width, color = Species)) + geom_point( ) + scale_colour_viridis_d(option = "plasma")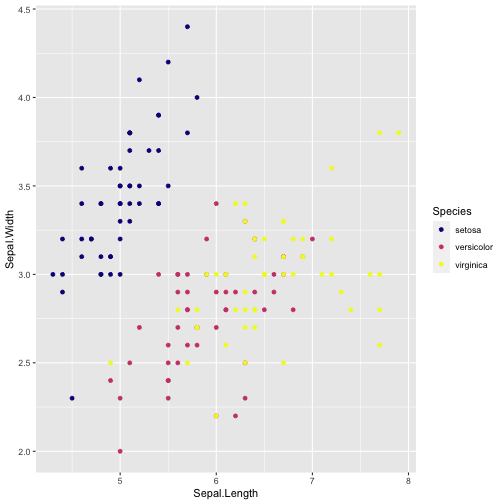
20 / 39
ggplot(iris, aes(x = Sepal.Length, y = Sepal.Width, color = Species)) + geom_point( ) + scale_colour_viridis_d(option = "inferno")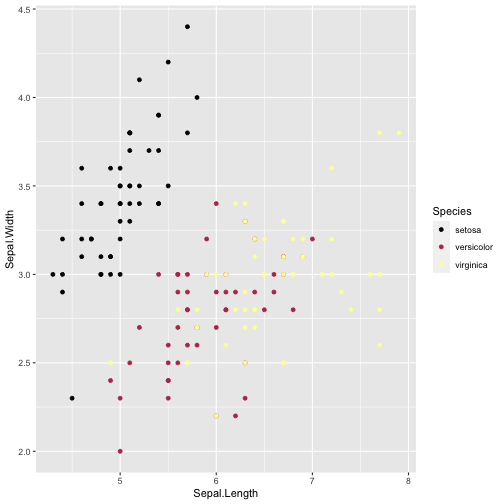
21 / 39
ggplot(iris) + geom_point(aes(x = Sepal.Length, y = Sepal.Width, color = Species, shape = Species, alpha = Species)) + scale_x_continuous( breaks = c(170,200,230)) + scale_y_log10() + scale_colour_viridis_d() + scale_shape_manual( values = c(17,18,19)) + scale_alpha_manual( values = c( "setosa" = 0.6, "versicolor" = 0.5, # "virginica" = 0.7))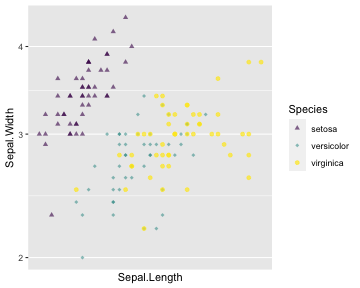
22 / 39
24 / 39
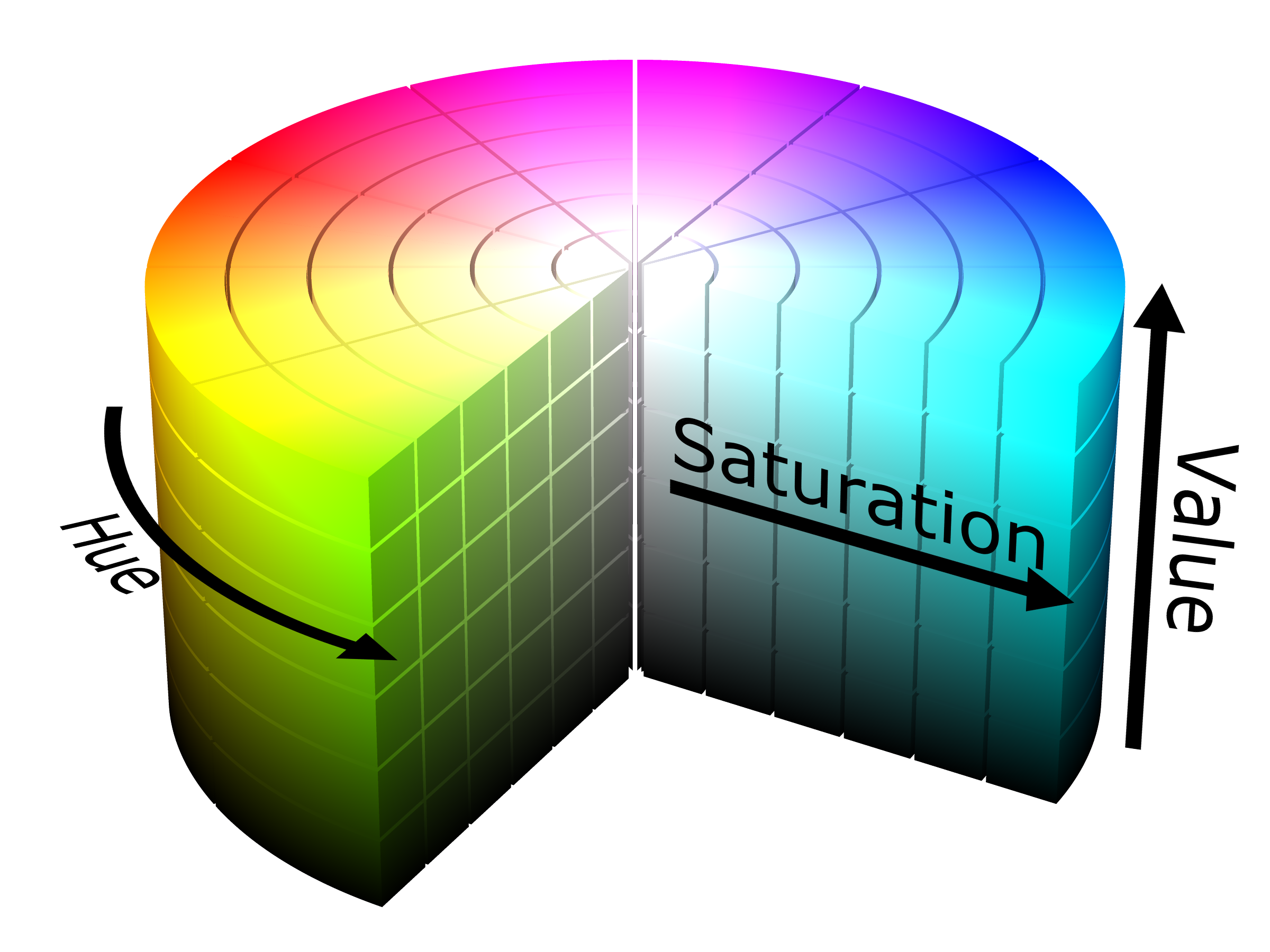
HSL colour attributes

Hue: true colours without tint or shades
Saturation: light tints to dark shades
Lightness (Value)
image source: https://purple11.com/basics/hue-saturation-lightness/
25 / 39
Coordinates
26 / 39
library(palmerpenguins)data(penguins)ggplot(penguins) + geom_bar(aes(x= species, fill = species))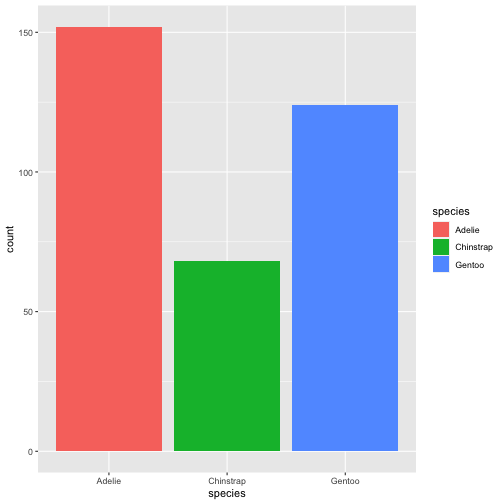
27 / 39
ggplot(penguins) + geom_bar(aes(x= species, fill = species)) + coord_flip()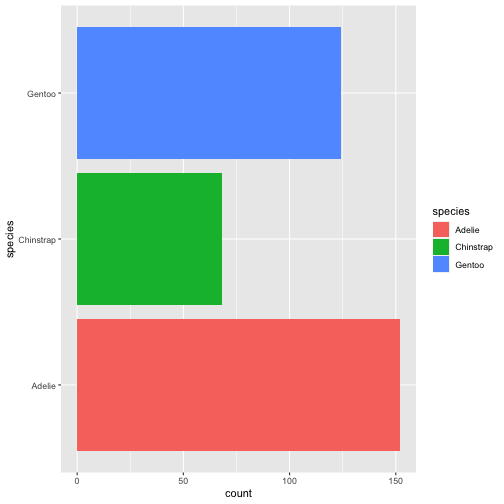
28 / 39
ggplot(penguins) + geom_bar(aes(x= year, fill = species))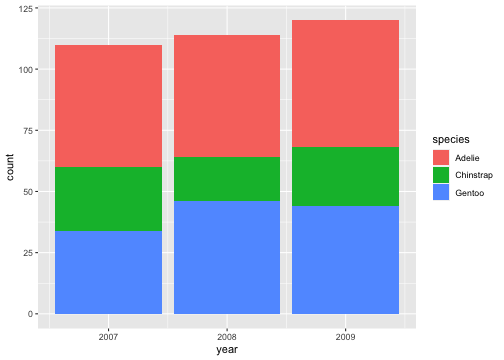
29 / 39
## Zooming into a plot with scaleggplot(penguins) + geom_bar(aes(x= year, fill = species)) + scale_y_continuous(limits = c(0,115))## Warning: Removed 1 rows containing missing values (`geom_bar()`).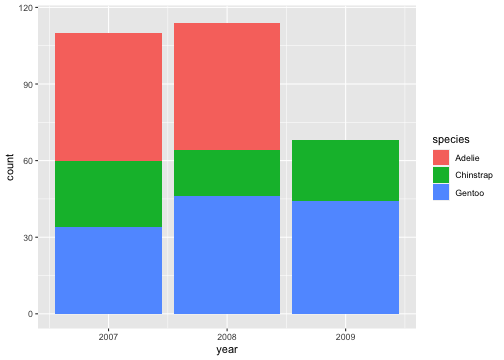
When zooming with scale, any data outside the limits is thrown away
30 / 39
Zooming into a plot with scale
ggplot(penguins) + geom_bar(aes(x= year, fill = species)) + coord_cartesian(ylim = c(0,115))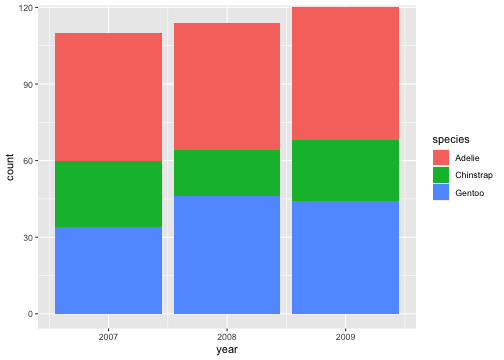
Proper zoom with coord_cartesian()
31 / 39
p <- ggplot(mtcars, aes(disp, wt)) + geom_point() + geom_smooth()p## `geom_smooth()` using method = 'loess' and formula = 'y ~ x'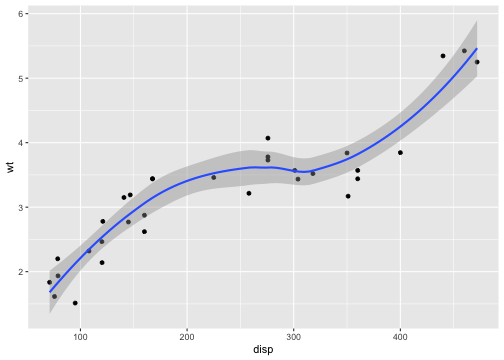
# Setting the limits on a scale converts all values outside the range to NA.p + scale_x_continuous(limits = c(325, 500))## `geom_smooth()` using method = 'loess' and formula = 'y ~ x'## Warning: Removed 24 rows containing non-finite values (`stat_smooth()`).## Warning: Removed 24 rows containing missing values (`geom_point()`).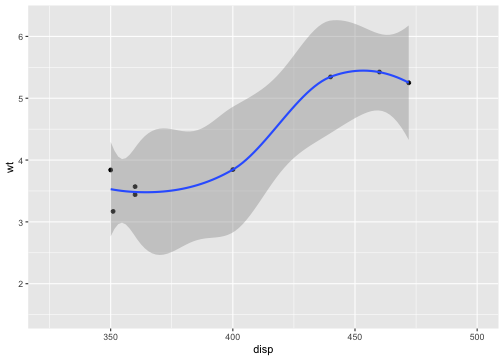
32 / 39
p <- ggplot(mtcars, aes(disp, wt)) + geom_point() + geom_smooth()p## `geom_smooth()` using method = 'loess' and formula = 'y ~ x'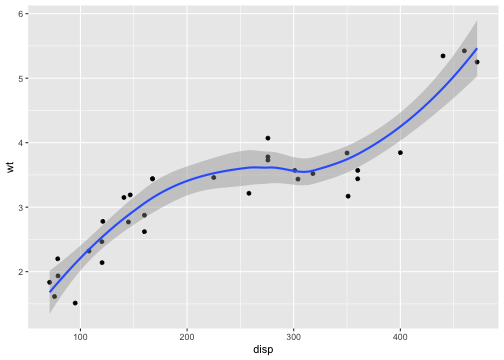
# Setting the limits on the coordinate system performs a visual zoom.# The data is unchanged, and we just view a small portion of the original# plot. Note how smooth continues past the points visible on this plot.p + coord_cartesian(xlim = c(325, 500))## `geom_smooth()` using method = 'loess' and formula = 'y ~ x'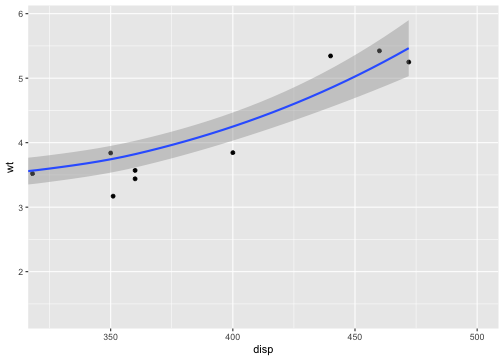
33 / 39
ggplot(iris, aes(y=Sepal.Length, x=Sepal.Width)) + geom_point()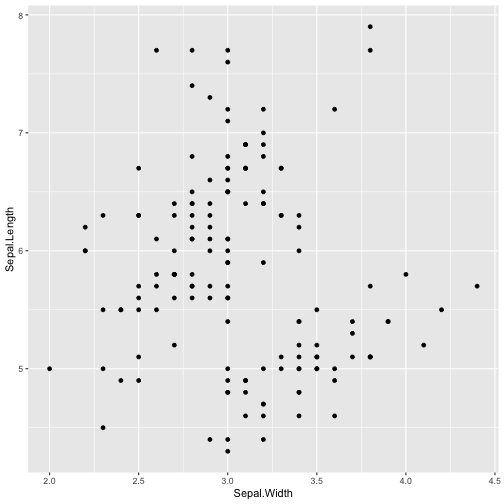
34 / 39
ggplot(iris, aes(y=Sepal.Length, x=Sepal.Width)) + geom_point() + coord_fixed(ratio = 1)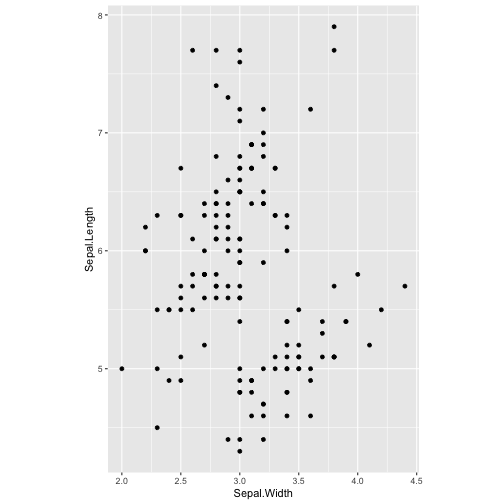
35 / 39
ggplot(iris, aes(y=Sepal.Length, x=Sepal.Width)) + geom_point() + coord_fixed(ratio = 0.5)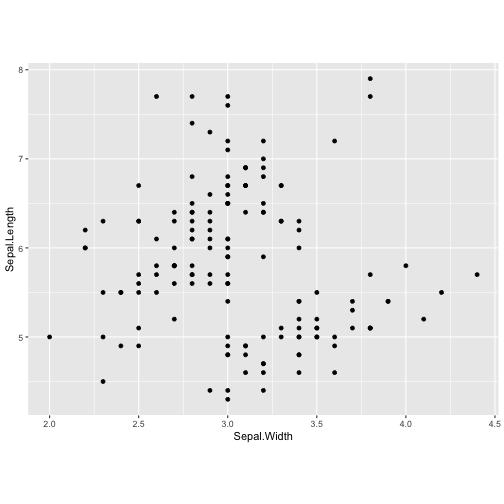
36 / 39
ggplot(iris, aes(y=Sepal.Length, x=Sepal.Width)) + geom_point() + coord_fixed(ratio = 5)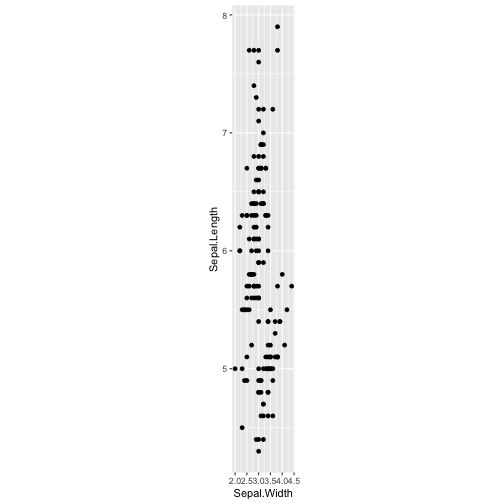
37 / 39
ggplot(iris, aes(y=Sepal.Length, x=Sepal.Width)) + geom_point() + coord_fixed(ratio = 1/5)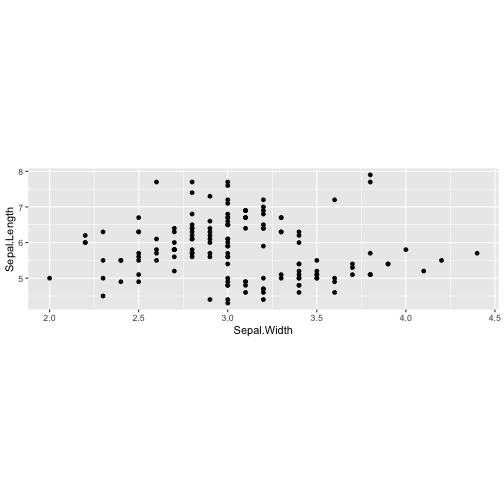
38 / 39
ggplot(iris, aes(y=Sepal.Length, x=Sepal.Width)) + geom_point() + coord_fixed(ylim = c(0, 6))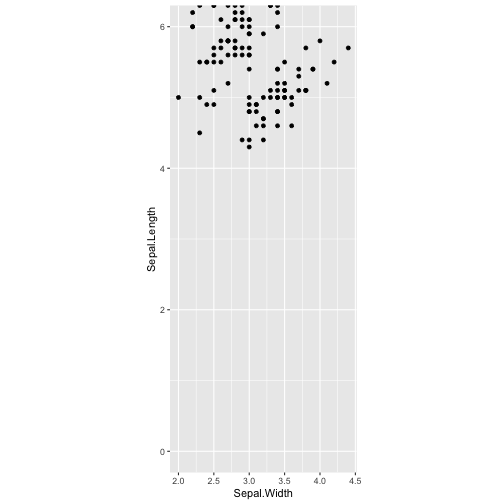
39 / 39