ASP 460 2.0 Data Visualisation
Visualizing Distributions
Thiyanga Talagala
1 / 40
Visualizing a Single Distribution
Histogram
Density plot
Cumulative density
Quantile-Quantile plot
Cumulative density and Quantile-Quantile plot are hard to interpret.
2 / 40
Visualizing multiple distributions
Visualization of distributions along the X-axis
Boxplots
Violins
Strip charts
Sina plots
Visualization of distributions at the same time
Staked histograms
Overlapping densities
Ridgeline plot
3 / 40
Histogram - Binwidth
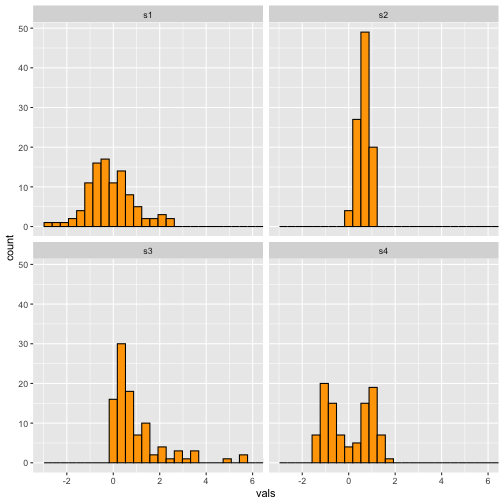
4 / 40
Histogram-Binwidth (.1)
Narrow
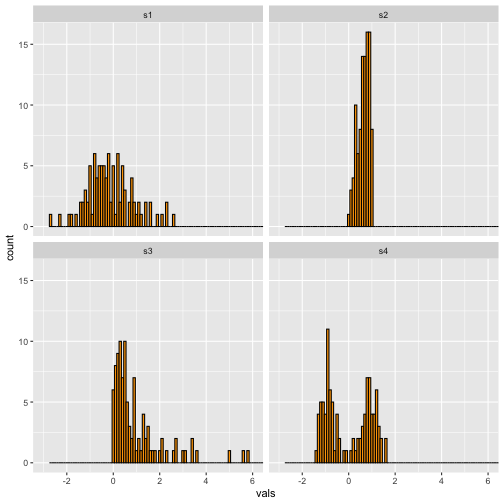
5 / 40
Histogram-Binwidth (2)
Wide
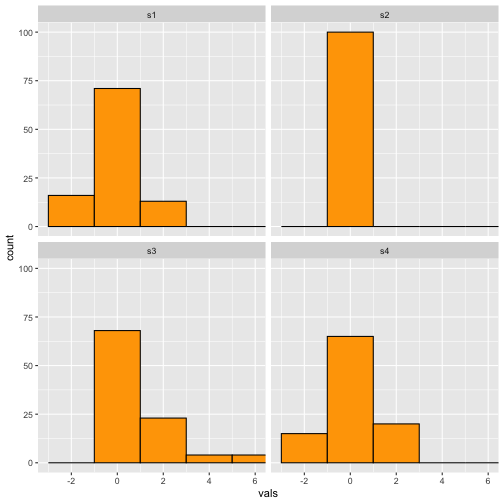
6 / 40
Add a rug
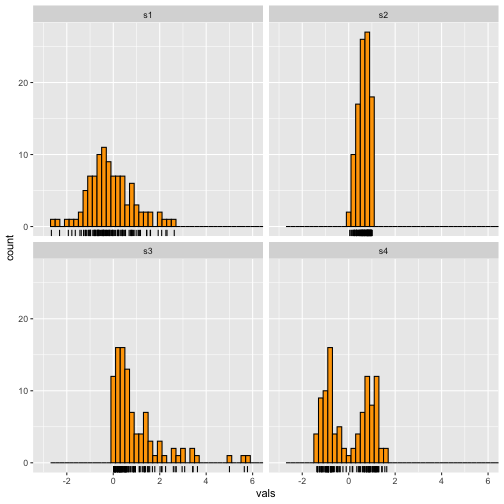
7 / 40
Histogram - Example
ggplot(iris, aes(x = Sepal.Length)) +geom_histogram(binwidth = .2, fill = "orange", colour = "black") +geom_rug() +facet_wrap(~ Species)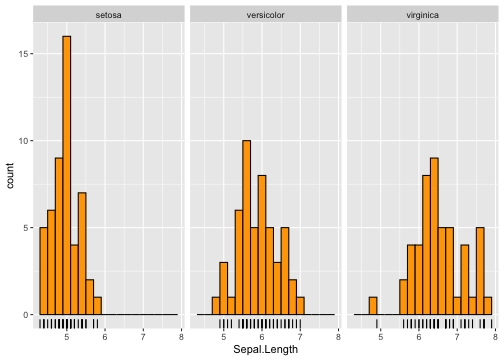
8 / 40
Boxplot
Medium to Large N
9 / 40
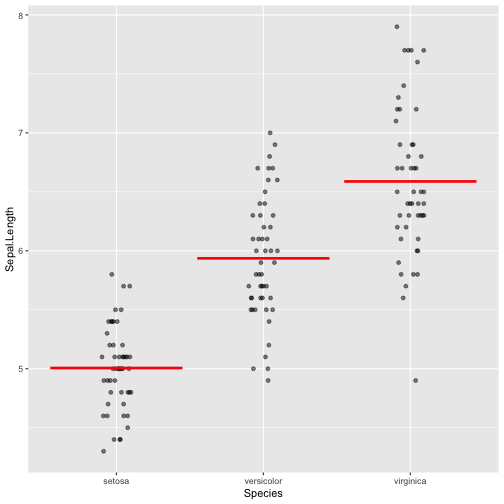
Boxplot - Example
ggplot(iris, aes(y = Sepal.Length, x = Species)) +geom_boxplot()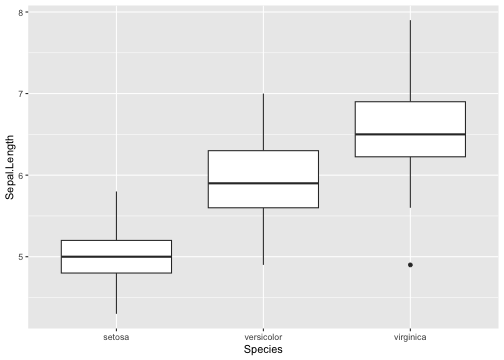
10 / 40
Add notches
“Notches are used to compare groups; if the notches of two boxes do not overlap, this is strong evidence that the medians differ.” (Chambers et al., 1983, p. 62)
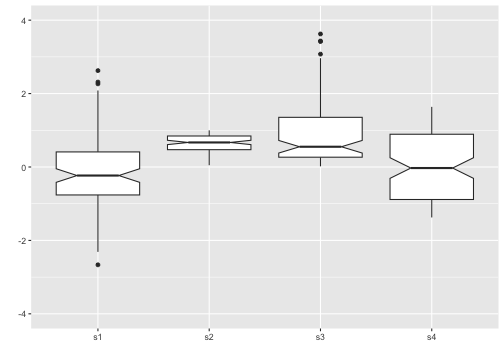
11 / 40
Boxplot with notch - Example
ggplot(iris, aes(y = Sepal.Length, x = Species)) +geom_boxplot(notch = T)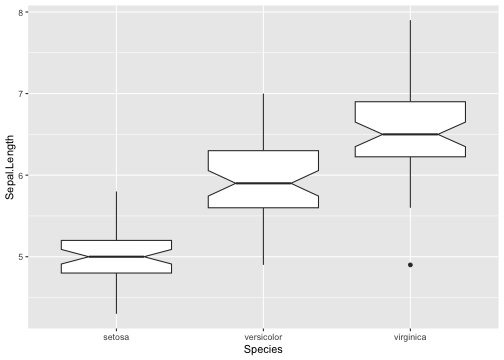
Your turn: Perform ANOVA.
12 / 40
Add summary statistics
Green: Mean
13 / 40
Boxplot with summary - Example
ggplot(iris, aes(y = Sepal.Length, x = Species)) +geom_boxplot() + stat_summary(fun.y=mean)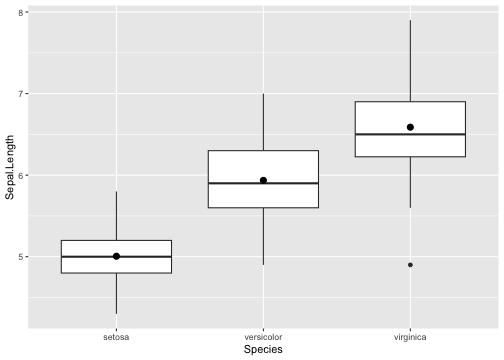
14 / 40
Boxplot with summary - Example
Your turn: Add min, max, Q1, Q2, Q3
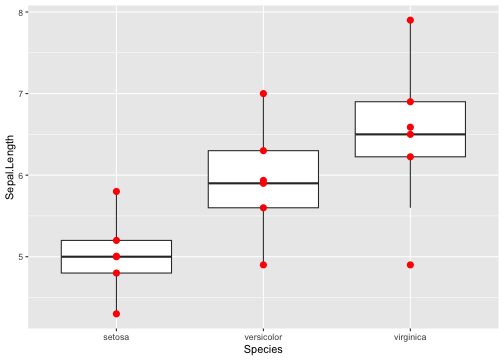
15 / 40
Stripchart
Small to Medium
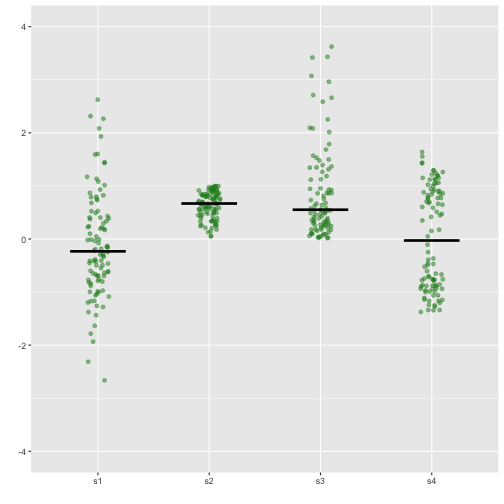
16 / 40
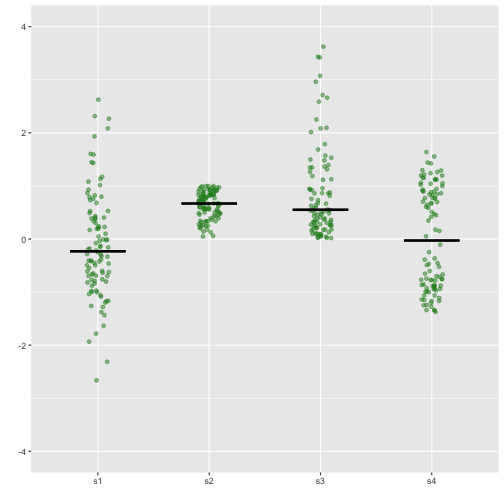
Stripchart - Example
17 / 40
Boxplot using geom_dotplot
Small to Medium
Previous
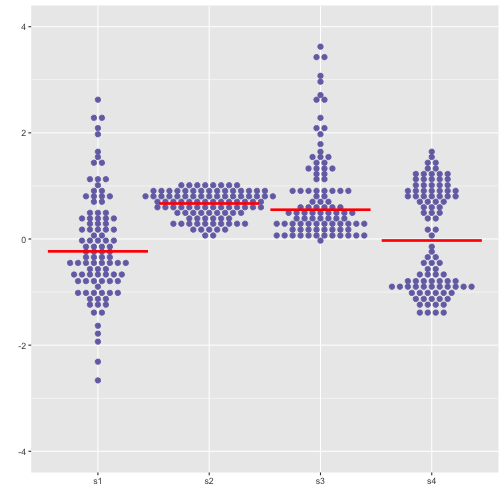
Now
18 / 40
Boxplot using geom_dotplot - Example
ggplot(iris, aes(x = Species, y = Sepal.Length)) +geom_dotplot(stackdir = "center",binaxis = "y", binwidth = .1,binpositions = "all",stackratio = 1.5,fill = "#7570b3", colour = "#7570b3")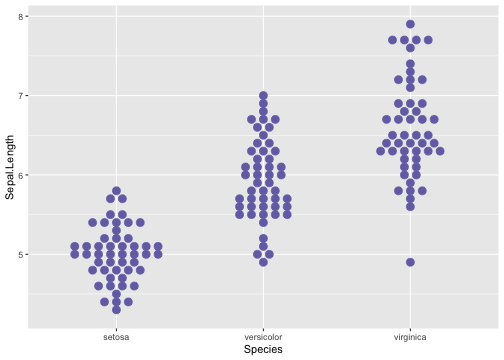
ggplot(iris, aes(x = Species, y = Sepal.Length)) +geom_dotplot(stackdir = "center",binaxis = "y", binwidth = .05,binpositions = "all",stackratio = 1.5,fill = "#7570b3", colour = "#7570b3")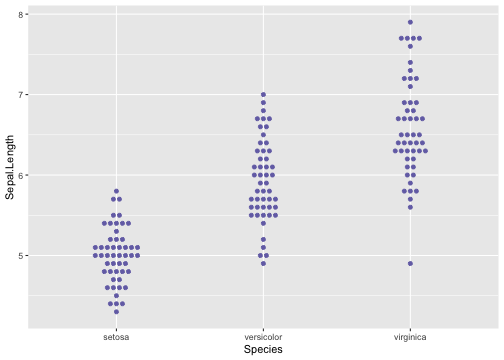
19 / 40
Bee swarm

20 / 40
Beeswarm
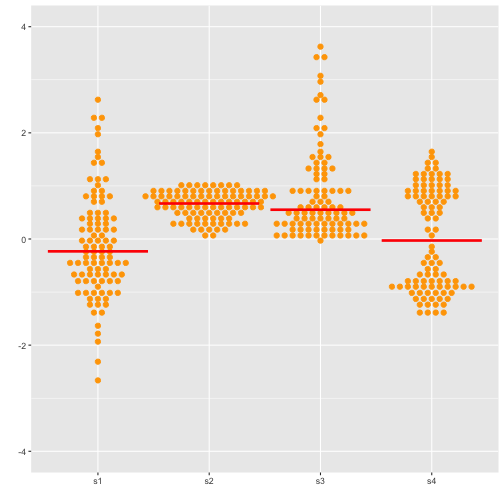
Previous
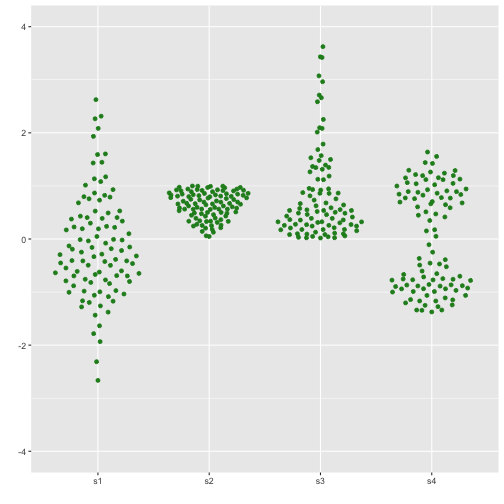
Now
21 / 40
Boxplot with dot points
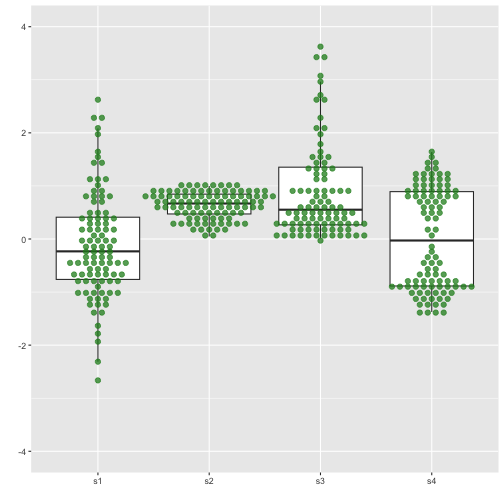
22 / 40
Boxplot with dot points - Example
ggplot(iris, aes(y = Sepal.Length, x = Species)) +geom_boxplot(outlier.shape = NA) +geom_dotplot(binaxis = 'y',stackdir = 'center', fill = "#7570b3", colour = "#7570b3", binwidth = .05)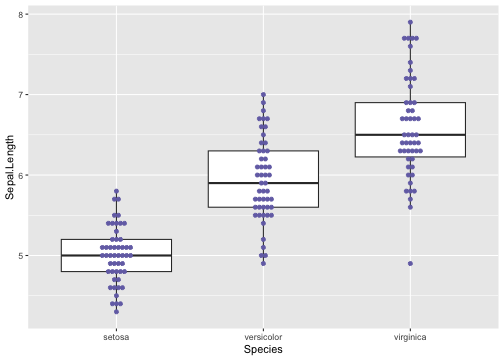
23 / 40
Boxplot with dot points
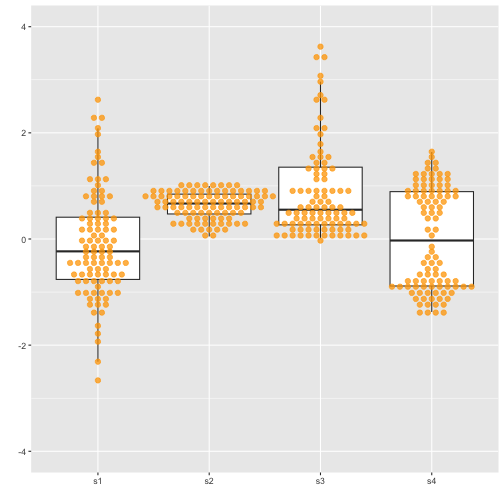
Previous
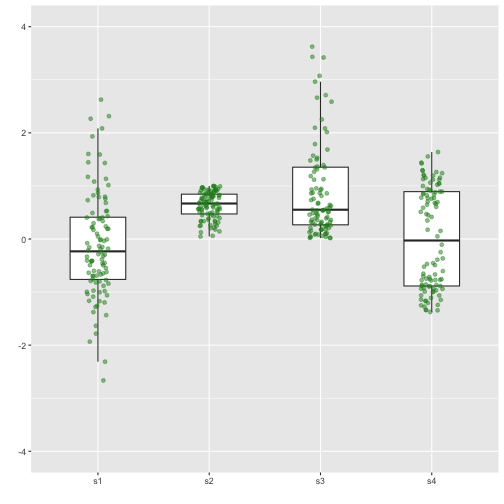
Now
with geom="jitter"
24 / 40
Boxplot with dot points (geom="jitter")
ggplot(iris, aes(y = Sepal.Length, x = Species)) +geom_boxplot(outlier.shape = NA, width = .5) +geom_jitter(fill = "#7570b3", colour = "#7570b3",position = position_jitter(height = 0, width = .1), alpha = .5)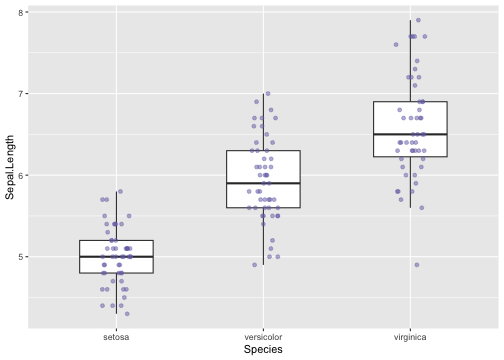
25 / 40
Density plots
Medium to large n
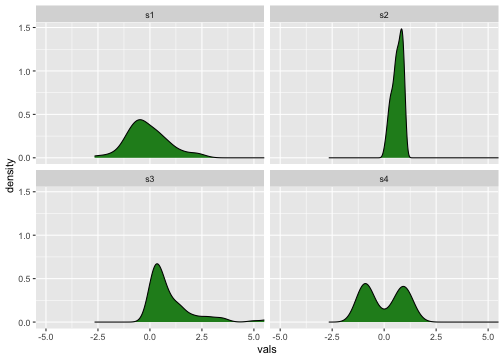
26 / 40
27 / 40
28 / 40
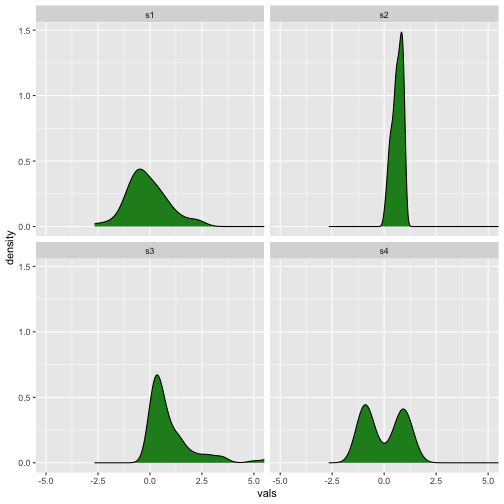
Density plot
Previous
Now
29 / 40
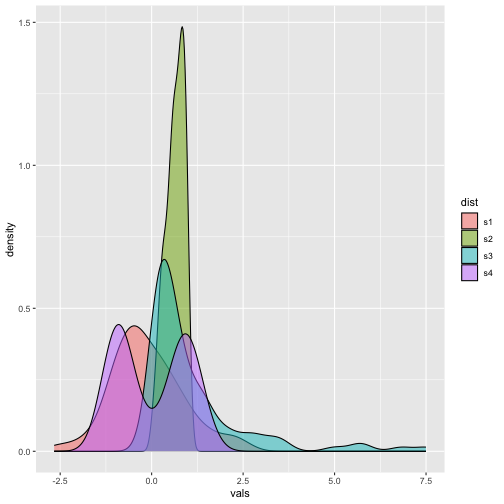
Density plots - Example
ggplot(iris, aes(x = Sepal.Length)) + geom_density(fill = "#7570b3") + facet_wrap(~ Species)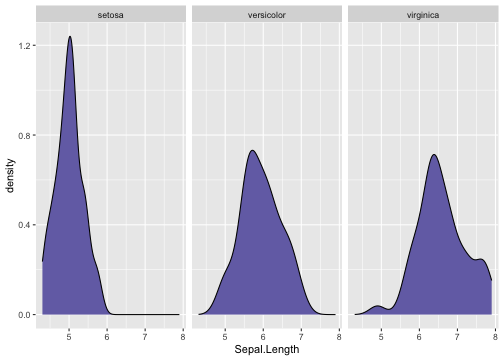
Previous
ggplot(iris, aes(x = Sepal.Length, fill=Species)) + geom_density(alpha=0.5)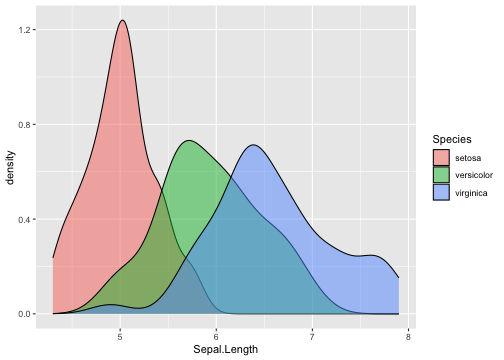
Now
30 / 40
Density plot and Histogram
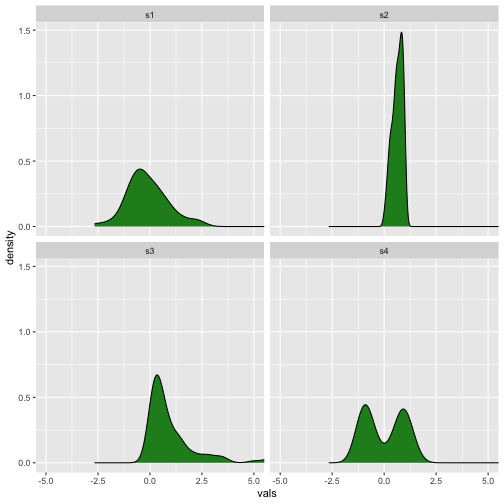
Previous
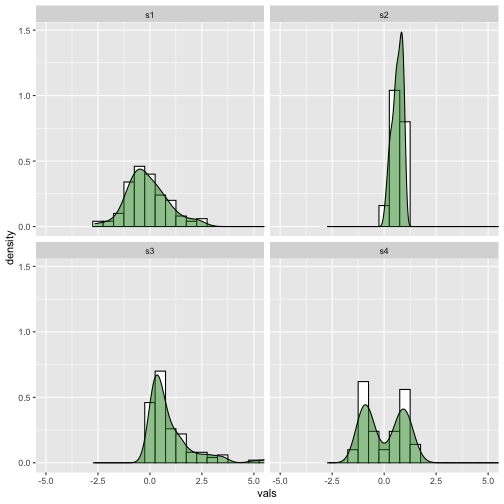
Now
31 / 40
Density plot and Histogram - Example
ggplot(iris, aes(x = Sepal.Length)) +geom_histogram(aes(y = ..density..),binwidth = .5, colour = "black",fill = "white") +geom_density(alpha = .5, fill = "#7570b3") +facet_wrap(~ Species)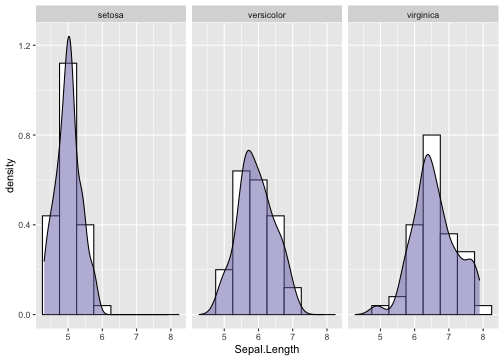
32 / 40
Violin plot
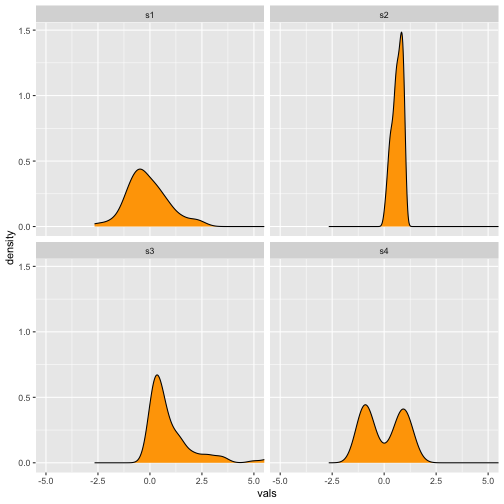
Previous
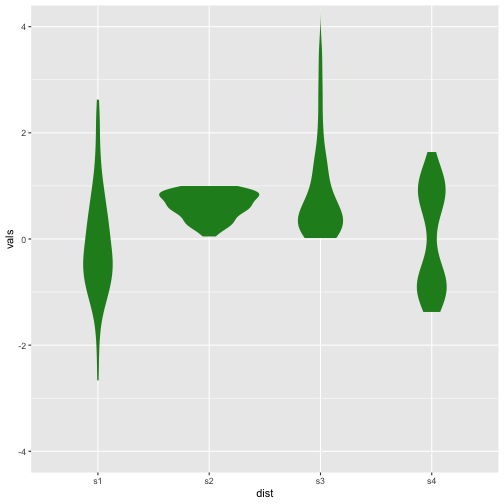
Now
33 / 40
Violin plot - Example
ggplot(iris, aes(x = Species, y = Sepal.Length)) +geom_violin(color = NA,fill = "#7570b3", na.rm = TRUE,scale = "count")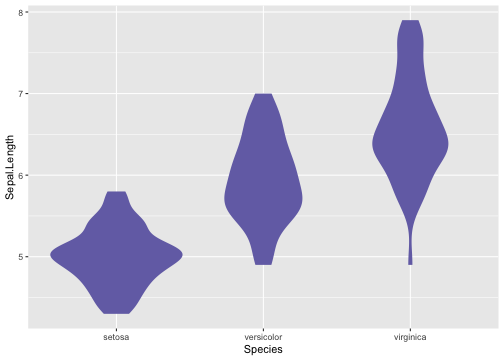
34 / 40
Violin plot + Boxplot
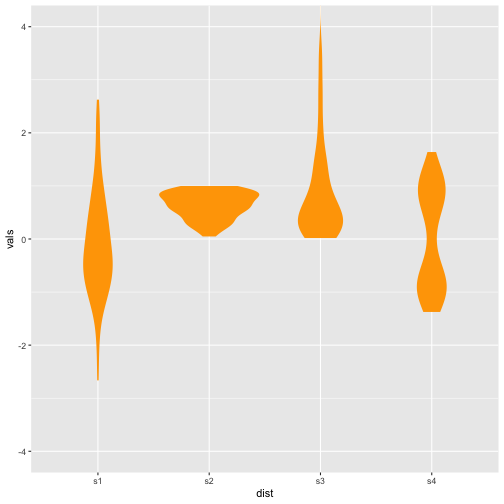
Previous
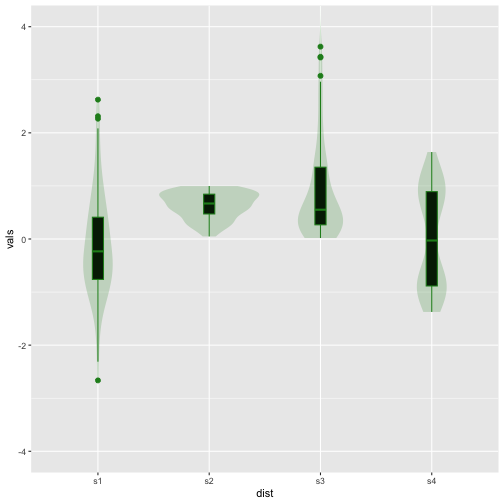
Now
35 / 40
Violin plot + Boxplot
ggplot(iris, aes(x = Species, y = Sepal.Length)) +geom_boxplot(outlier.size = 2, colour="#7570b3", width=.1) +geom_violin(alpha = .2, fill = "#7570b3")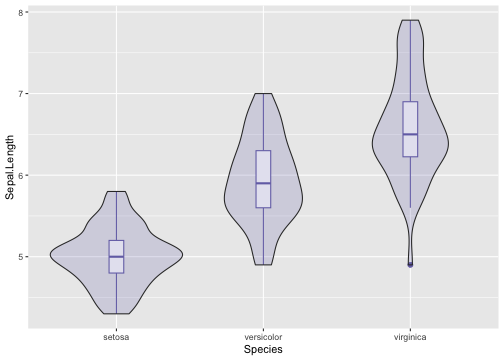
36 / 40
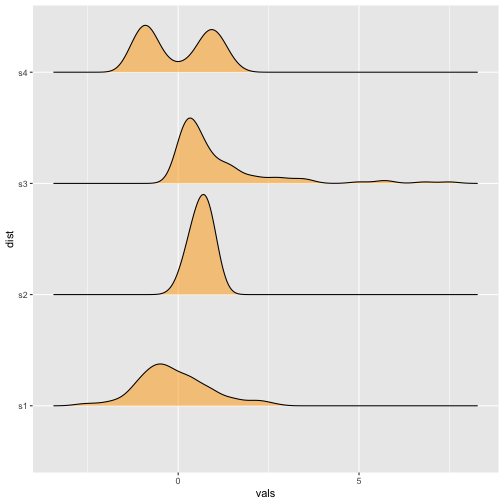
Ridgeline plots
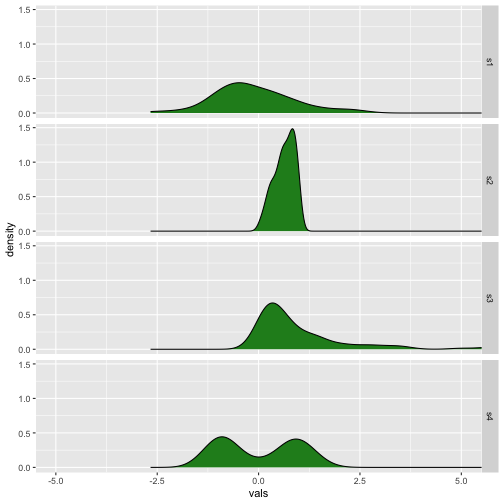
Previous
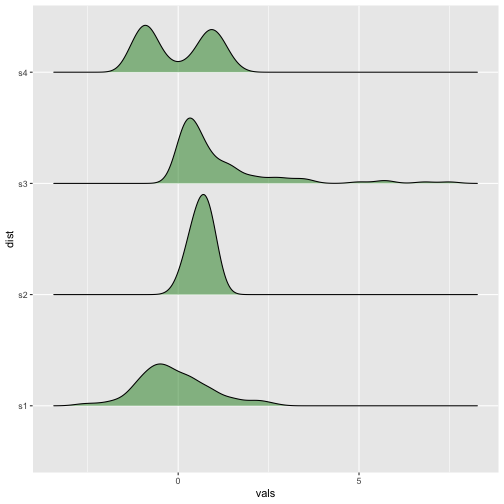
Now
37 / 40
Ridgeline plots - Example
library(ggridges)ggplot(iris, aes(x = Sepal.Length, y = Species)) +geom_density_ridges(scale = 0.9,fill = "#7570b3", alpha = .5)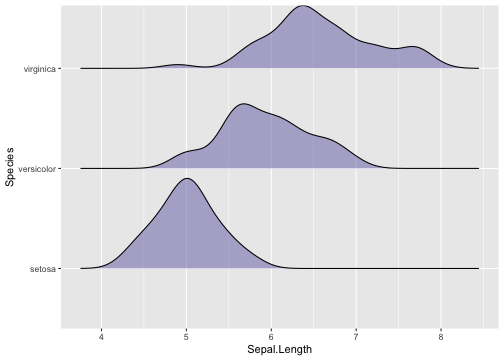
38 / 40
Raincloud plot
Previous
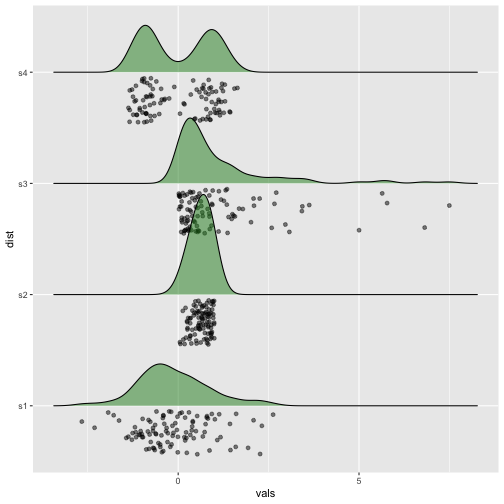
Now
39 / 40
Raincloud plots - Example
library(ggridges)ggplot(iris, aes(x = Sepal.Length, y = Species)) +geom_density_ridges(scale = 0.9, position= "raincloud", jittered_points = TRUE,fill = "#7570b3", alpha = .5)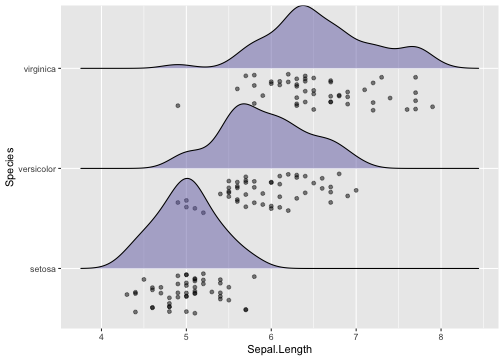
40 / 40